Abstract
A paradigm shift in drug discovery and development has been spurred on through advances in the emerging field of synthetic biology. This article discusses the potential of synthetic biology to transform several stages of the drug discovery and development process and enhance the drug pipeline. The impact would be critical for the dawn of a brave new world for the pharmaceutical industry.
The success of the Pharma industry is heavily incumbent on the quality and quantity of drugs in the pipeline. The urgent imperative for new drivers of pharmaceutical drug discovery and development are implicit in the declining pipeline of drugs approved annually by the FDA and the increase in drug resistance world-wide. The process of drug discovery is complex and time intensive ranging from years to decades, and success at each phase of development is unpredictable. In the last century, synthetic organic chemistry was the centre of innovation in the pharma industry. Challenges to succeed include but are not limited to animal model validity, unknown disease mechanisms, patient heterogeneity, poor targets, weak biomarkers, clinical trial outcomes and regulatory challenges [1]. In this era, the new emerging discipline of synthetic biology has the potential to completely reorient and revolutionize the pharmaceutical industry. The potential increase in drugs at the clinical stage reaching the market will be a huge benefit. Even greater will be the ability to drive personalized medicine and individualized therapy by ensuring efficacy in target populations.
In contrast to Paul Ehrlich’s one drug-one target paradigm [2], chemical proteomics data has unravelled multiple targets for a specific drug. Thus, it is no longer sufficient to envision small molecule ligand interactions alone, but one has to understand poly-pharmacology, cross-talk and complex, adaptive molecular networks in humans and across populations. The advent of genome-scale measurement technologies has spurred on holistic thinking and systems approaches in biology and medicine. In order to harness unprecedented opportunities for drug discovery, rational therapeutics and personalized medicine are central. There is an obligate need for a paradigm shift in thinking and systems medicine and synthetic biology are critical drivers shaping this process. The emerging applications of synthetic biology to pharmaceutical industry for drug discovery, and development are relatively new and hold great promise. Synthetic biology startups and hackers in their garages and basements are using cloud-based services, do-it-yourself manuals along with low-cost nucleotide sequence construction, laboratory tools, and equipment to manipulate organisms to produce materials and products of economic value.
Drug Discovery and Synthetic Biology
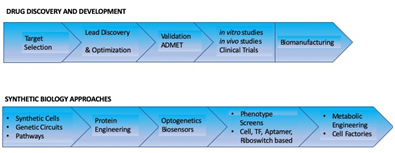 Synthetic and systems biology can be used at different stages of the drug discovery process including preclinical, clinical and manufacturing(Figure 1). This could involve prediction and validation for target identity, selection, screening and hit to lead discovery and optimization; efficacy and safety testing in clinical trials and biomanufacturing.
Synthetic and systems biology can be used at different stages of the drug discovery process including preclinical, clinical and manufacturing(Figure 1). This could involve prediction and validation for target identity, selection, screening and hit to lead discovery and optimization; efficacy and safety testing in clinical trials and biomanufacturing.
Accurate drug target identification and validation using standard knock-out and knock-in experiments in cell-lines and/or mice are critical to successful drug development process. Synthetic cells can now be used to validate targets and also further identify drug–target interactions by using small molecules to modulate expression of essential gene targets [3]. Construction of disease-related genetic circuits within minimal synthetic cells promoting cell survival can be used for screening during hit to lead optimization. Phenotypic screening can be based on synthetic cells or transcription factors (TFs), aptamers, riboswitches or ribozymes. Cell-based phenotypic assays have the ability to predict the impact of the drug on an entire pathway in contrast to a single component. Cross talk between pathways across molecular hierarchies leading to imbalance or toxic side products can be delineated.
Clinical trials can be reduced through predictive modeling and lab-on-a-chip technologies. Synthetic cellular models of disease in pharmaceutical research can double up as diagnostics or therapeutic strategies to decipher drug response and pathogenesis. Disease models can be typically engineered through genetic circuits to study cellular responses by controlling molecular mechanisms through light stimuli (optogenetics). Luminescent or fluorescent reporter synthetic gene circuits can record the multifactorial response of potential biomarkers to varying physiological conditions like UV stress, growth factors and drugs. Synergies between pharmacogenetics, chemo-genetics and optogenetics can also be leveraged at every stage. Drug-resistance and persistence mechanisms can be probed using synthetic quorum sensing networks simulating cell-cell communication within bacterial consortia and biofilms. Artificial communication systems can be engineered to identify compounds that disrupt quorum sensing for drug resistance.
Biomanufacturing holds great promise for pharmaceutical production, however, the number of products at industrial scale (50 g/L) are still limited to a few amino acids and isoprenoids, while medium scale (5–50 g/L) is feasible only for artemisinin and specific antibiotics [1]. Synthetic cells with large biosynthetic gene clusters or modules are cell factories for desired products. Genetic shuffling of biosynthetic modules allows for novel modifications. One of the biggest success stories of synthetic biology is in the area of biomanufacturing. The production of Amorpha-4, 11-diene in yeast, and its conversion to dihydroartemisinic acid, precursor to the antimalarial agent artemisinin [4] has been a massive effort. A modified mevalonate pathway was designed and implemented in yeast cells to express the enzyme amorphadiene synthase and a cytochrome P450 monooxygenase (CYP71AV1) both from Artemisia annua. A three-step oxidation of amorpha-4,11-diene resulted in artemisinic acid. Other products being optimized commercially through this route include 1,4-Butanediol and 1,3 Propane Diol by Genomatica Inc., and Dupont, Inc. respectively [5].
Biomanufacturingand Synthetic Biology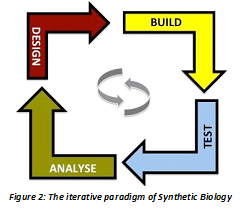 Synthetic biology may be looked upon as an interdisciplinary branch at the interface of biology and engineering that allows the application of engineering and design principles to build and optimize cellular processes in organisms. In biomanufacturing, there is an implicit view of the biological cell as the ultimate chemical factory. Analogous to unit operations, connectors and process controllers in a chemical factory, the component classes in cells are represented as organelles, genes, proteins, regulators. However, there is a need to refine or fine-tune them to achieve metabolic control at the level of sophistication in a chemical plant. Harnessing the modularity and hierarchy existent in biological systems allows fabrication of such plug and play models for biosynthetic clusters of varying complexity. Thus one can create designer organisms/strains through rational design that efficiently converts different carbon sources into a diverse range of products, making the process sustainable with the promise of viable economic cost. This process allows access to 3-tiers of optimization: the central dogma, intrinsic regulatory interactions and extrinsic environmental interactions leading to their seamless integration. The development of rational strain designs through the Design-Build-Test-Analyze (Figure 2) paradigm of synthetic biology [6] would fast forward evolution and create scalable workflows for optimization of drugs and other value-added products.
Synthetic biology may be looked upon as an interdisciplinary branch at the interface of biology and engineering that allows the application of engineering and design principles to build and optimize cellular processes in organisms. In biomanufacturing, there is an implicit view of the biological cell as the ultimate chemical factory. Analogous to unit operations, connectors and process controllers in a chemical factory, the component classes in cells are represented as organelles, genes, proteins, regulators. However, there is a need to refine or fine-tune them to achieve metabolic control at the level of sophistication in a chemical plant. Harnessing the modularity and hierarchy existent in biological systems allows fabrication of such plug and play models for biosynthetic clusters of varying complexity. Thus one can create designer organisms/strains through rational design that efficiently converts different carbon sources into a diverse range of products, making the process sustainable with the promise of viable economic cost. This process allows access to 3-tiers of optimization: the central dogma, intrinsic regulatory interactions and extrinsic environmental interactions leading to their seamless integration. The development of rational strain designs through the Design-Build-Test-Analyze (Figure 2) paradigm of synthetic biology [6] would fast forward evolution and create scalable workflows for optimization of drugs and other value-added products.
Rational construction of desired metabolic traits in a heterologous host began several decades ago and has now progressed to the assembly of very complex pathways. One of the reasons for major success of synthetic biology in biomanufacturing is due to the plethora of techniques developed over the last 2-3 decades in the sister sciences of metabolic engineering and systems biology. Thus, the synergy between synthetic biology, systems biology and metabolic engineering has potentially lead to microbial cell factories(Figure 3). Genetically well characterized hosts like E. coli and S. cerevisiae, with favourable growth rates and simple nutrient requirements are attractive biosynthetic hosts. In recent years, metabolic engineering has shifted its paradigm towards a systemic approach that relies on large scale experimentation and computational analysis of metabolic and regulatory networks. High throughput technologies have resulted in databases that contain biochemical, molecular and genomic information that enable more systemic metabolic engineering approaches. The reconstruction of in silico genome scale stoichiometric models of metabolic networks that compute the functional state of the cell accurately facilitate a priori determination of effects of gene deletions and insertions on metabolic state, growth and redox state for improved product synthesis [7].
It is possible to extend the spectrum of drugs synthesized to include novel domains. However, the complexity of metabolic pathways, along with regulation often results in unexpected outcomes for metabolic engineering and the resulting strains may require extensive fine tuning to be economically viable.
Systems Biology in Pharmaceutical Research
We are at the helm of a paradigm shift in designing new drugs and therapies. Systems thinking in the biology and medicine has led to the use of live cells and molecular components of cells for personalized medicine and individualized therapy. The central dogma of molecular biology, i.e, information stored in DNA is transcribed to mRNA and then translated into a protein that carries out catalytic, regulatory or structural function in a cell, governs cell function and gives it a hierarchical multi-scale structure. Thus, integrating transcript, protein and metabolite information is essential to understand the phenotype under specific diseased conditions. More recently, these techniques have been exploited for design purposes and engineering a cell for a desired trait or function forming the basis of gene therapy and tailoring microenvironments in the cell. Metabolic 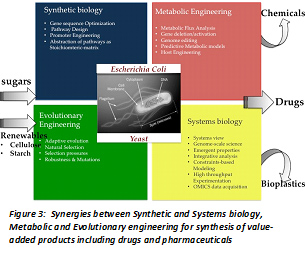 engineering has benefited immensely by large scale screening and implementation, due to the global effects of local changes in pathways to maximize a desired product. Further, building in silico models (computational and mathematical) has allowed us to analyze complex network structure to ensure stability, robustness to perturbations and display emergent properties for manipulation.
engineering has benefited immensely by large scale screening and implementation, due to the global effects of local changes in pathways to maximize a desired product. Further, building in silico models (computational and mathematical) has allowed us to analyze complex network structure to ensure stability, robustness to perturbations and display emergent properties for manipulation.
Challenges for Synthetic Biology in Pharmaceutical Industry
The grand challenges of the pharmaceutical industry are evident in the complexity of cellular pathways and pleiotropic effects of molecular components in biological systems and their dynamic states. The marriage of reductionist understanding of pathways in the cell and integrative knowledge of how these pathways are networked are critical in design of virtual cells that have predictive value. There has been a shift in the classes of drugs being developed from small molecules to biologics. Novel therapies that involve gene editing have become a reality for patients suffering from genetic conditions. CRISPR (Clustered, Regularly Interspaced, Short Palindromic Repeats) is a naturally-occurring defence mechanism used by bacteria that has now been harnessed to develop ‘molecular scissors’ that can remove mutated areas of DNA in certain diseases. This is hugely controversial because it changes the fundamental genetic code inherited by an offspring.
Cells with minimal genome and faster growth rate is the key to the success of any strategy, so the key is to find the minimum set of genes required to sustain life [8]. As construction of genetic circuits is key to screening and drug-target interaction validation, the assembly of standard parts is another area that needs a constant improvement. More recombination compatible systems are necessary to make strategies to clone genes. In rational strain design, a major limitation is plasmid instability in the cell because of metabolic burden to maintain and replicate plasmids, solution for this problem is the genome integration of desired construct that has high stability. Some limitations that need to be overcome for high performance in biomanufacturing include cofactor and energy imbalance, toxicity, enzyme activity, metabolic burden can be addressed using pathway engineering that exploits the synergies of synthetic biology, metabolic engineering and systems biology [7]. Complementation of nutrient precursors and cofactors that support coupling of cellular objectives of growth and energy to desired bioengineering objectives are necessary and have been identified through pathway analysis [9].
Security and Risk in Synthetic Biology
Understanding the risk involved in this field is critical. It is under the tight regulation by security agencies across the world, it is important to share the details of every gene that is synthesized in any corner of the world to stakeholders of world security(IGSG, 2013). Most of the synthetic biology projects use laboratory strains that cannot survive in the wild which prevent the potential environmental hazards from engineered strains. It should be taken care that synthetic genes/artificial gene circuits should not escape the boundaries of laboratory.
Future directions
The design principles and engineering tools to build and optimize cellular processes are state of the art today and encompass the field of synthetic biology. This process allows access to 3tiers of optimization: the central dogma, intrinsic regulatory interactions and extrinsic environmental interactions leading to their seamless integration. Synthetic biology lends itself elegantly to metabolic engineering and microbial cell factories and can be customized to the pharmaceutical industry. However, the complexity of metabolic pathways, along with regulation often results in unexpected outcomes for metabolic engineering and the resulting strains may require extensive fine-tuning to be economically viable.
Therapeutic solutions using synthetic mammalian cells to tailor microenvironments have only just begun and would potentially become the next generation cures for disease [10]. The holy grail would be to create a universal cell that can turn on biosynthesis of any drug selectively by specific sensed disease phenotypes. Lead optimization through selection pressures of microenvironments and resource allocation and choices. Varying microenvironment to modulate transcript expression and trigger molecular switches can be used potentially as a therapeutic strategy. We foresee workflows integrating computational and experimental strategies in systems metabolic engineering and synthetic biology as invaluable for generating strain portfolios for pharmaceutical drugs and intermediates and critical in realizing the potential of personalized medicine.
References:
[1] Trosset JY and Carbonel P, Synthetic biology for pharmaceutical drug discovery, Drug Design, Development and Therapy 2015:9, 6285-6302
[2] Mulinari S. The specificity triad: notions of disease and therapeutic specificity in biomedical reasoning. Philos Ethics Humanit Med. 2014;9:14.
[3] FirmanK, EvansL, YouellJA., Syntheticbiologyproject–developing a single-molecule device for screening drug-target interactions. FEBS Lett. 2012;586(15):2157–2163.
[4] Paddon CJ, Keasling JD. Semi-synthetic artemisinin: a model for the use of synthetic biology in pharmaceutical development. Nat Rev Microbiol. 2014;12(5):355–367
[5] Chubukov, V., Mukhopadhyay, A., Petzold, C.J., Keasling, J.D., Martín, H.G., 2016. Synthetic and systems biology for microbial production of commodity chemicals. npj Syst. Biol. Appl. 2, 16009
[6] Immanuel, S.R.C., Banerjee, D., Rajankar, M.P., Raghunathan, A., 2018. Integrated constraints based analysis of an engineered violacein pathway in Escherichia coli. Biosystems 171, 10–19.
[7] Nielsen, J., Keasling, J.D., 2011. Synergies between synthetic biology and metabolic engineering. Nat. Biotechnol. 29, 693–695.
[8] Hutchison, C.A., Chuang, R.-Y., Noskov, V.N., Assad-Garcia, N., Deerinck, T.J., Ellisman, M.H., Gill, J., Kannan, K., Karas, B.J., Ma, L., Pelletier, J.F., Qi, Z.-Q., Richter, R.A., Strychalski, E.A., Sun, L., Suzuki, Y., Tsvetanova, B., Wise, K.S., Smith, H.O., Glass, J.I., Merryman, C., Gibson, D.G., Venter, J.C., 2016. Design and synthesis of a minimal bacterial genome. Science (80) 351, 6253-6253
[9] Klamt, S., Mahadevan, R., 2015. On the feasibility of growth-coupled product synthesis in microbial strains. Metab. Eng. 30, 166–178.
[10] Lienert F, Lohmueller JJ, Garg A, Silver PA., Synthetic biology in mammalian cells: next generation research tools and therapeutics., Nat Rev Mol Cell Biol. 2014 Feb;15(2):95-107.